For researchers
Maximize your research potential with precision whole genome sequencing, curation, and interpretation, all in 2 weeks.

Our solutions
Our integrated and customizable whole genome solutions
Sequencing
In our CAP/CLIA lab with latest sequencing technology
Bioinformatics
With additional insights that only whole genome can bring
CAP/CLIA assays
For cancer and rare diseases, supporting FDA submission for Phase II and III clinical trials
Genomic datasets
With more than 13,000 whole genome data with various patient cohorts
We specialize in whole genome sequencing + analytics
Sequencing
- High quality services in a CLIA-certified, CAP-accredited lab
- With or without DNA extraction and library prep
- Latest sequencing technology – NovaSeqTM X Plus, Ultima UG 100TM
- Flexible raw data files (typically FASTQ, BAM, CRAM)
Bioinformatics
- World-class bioinformatics providing whole genome insights
- Analytics, interpretation, or/and report generation
- Curated data files (typically VCF) or Inocras Dx product reports
- Customized report available depending on volume
Sequencing + Bioinformatics
- We perform end-to-end services, including sequencing (with or without library prep) and bioinformatics (with or without report generation) — performed in 2 weeks
Sequencing only
Bioinformatics only
End-to-end services
Expert advisory support
Our team of MDs and PhDs specializes in medical, genomic, and computer science and bioinformatics, and provides advisory services on an hourly or project basis.
Powering the future of genomic research
Our research services are powered by revolutionary technology and backed by a team of world-class experts in medical, genomic science, computer science, and bioinformatics.
CLIA certified, CAP accredited
generating high quality whole genome data
Reliable 2 week turnaround time
powered by operational excellence in our CAP/CLIA lab
World-class bioinformatics
with proprietary pipeline and high-automation, enabling accuracy and consistency in curation and interpretation
End-to-end support
customized to meet your unique research needs and project goals
Leverage our premium and comprehensive solutions
Oncology platform
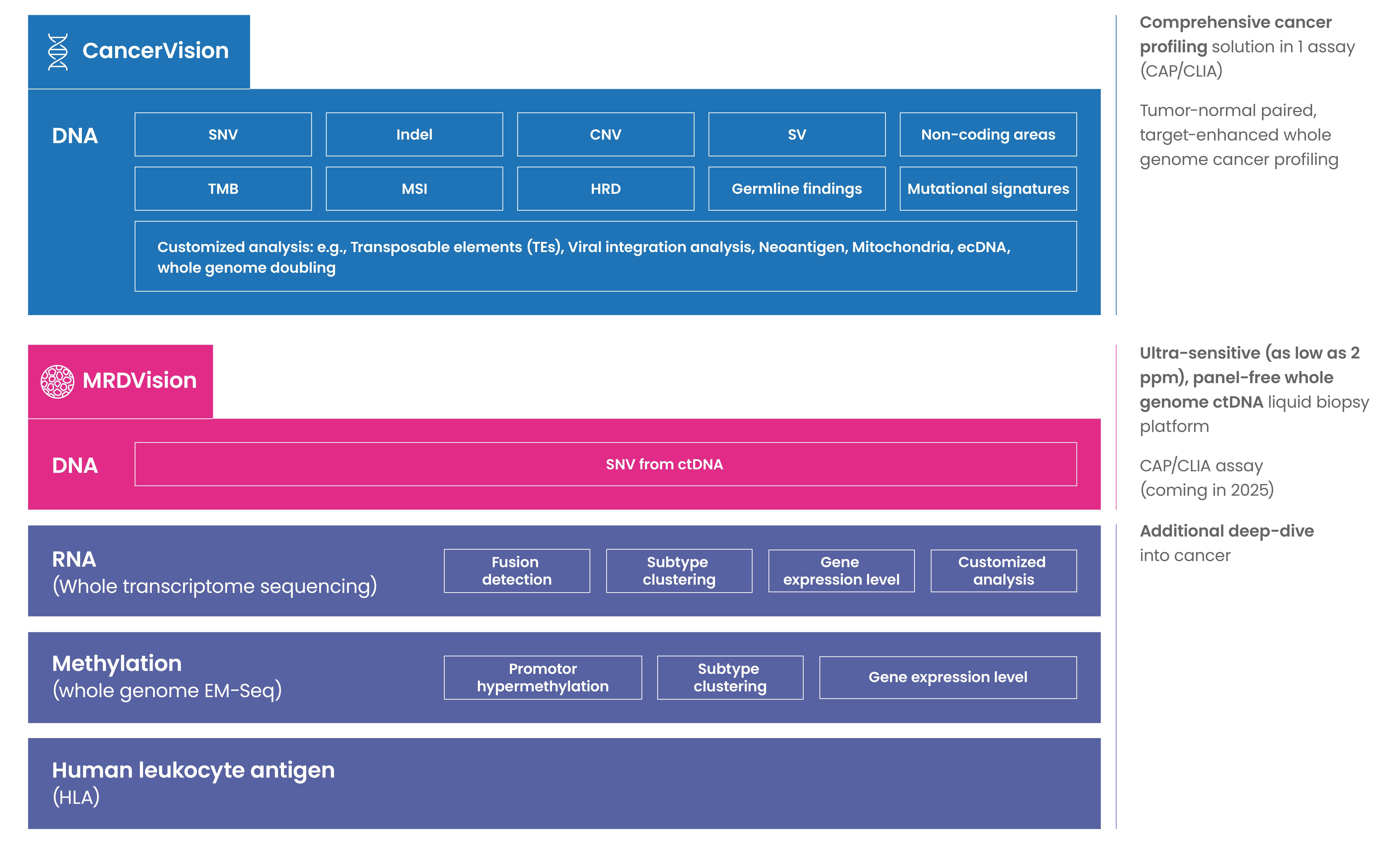
Germline platform
Our research services are customizable and tailored to your unique needs.
If you don’t see the service you’re looking for here, please connect with us.
Accelerate your research with whole genome insights
Biopharmaceuticals and biotechnology
- Identify candidate biomarkers that correlate with outcomes
- Understand responders vs. non-responders during or post clinical trials
- Accelerate clinical trial enrollment by identifying patients for rare indications
- Analyze clinical trial results for efficiency and safety profile
- Leverage genetic information for label expansion with real world data
- Provide genetic testing for patient support program
Biobanks, research organizations and labs
- Research genomic profiles in-depth, often resulting in new biomarker identification, or re-classification
- Profile genomic characteristics of the acquired biospecimen to increase the value of your biospecimen assets
Health technology
- Leverage genetic information to provide precision health insights to your customers and users
- Generate real world evidence data including genetic information
Sample requirements
Normal samples
- gDNA: ≥100ng, minimum concentration of 2ng/uL, shipped ambient or frozen
- Saliva: 1 tube in collection device, ambient
- Peripheral blood: ≥3mL of peripheral blood, EDTA or Streck tube, ambient
- Buccal swab in stabilization solution, ambient
- Adjacent normal tissue: ≥10 slides in 5 micron thickness, ambient
- Buffy coat: shipped frozen on dry ice
Tissue samples
- Fresh frozen tissue: 25mg of cryopreserved tissue, shipped frozen on dry ice
- FFPE tissue: ≥10 slides in 5 micron thickness, ambient